
The gastrointestinal microbiome and the enteropathogenetic syndromes: an infectious diseases perspective
Introduction
Human microbiota refers to all of the microbes that are present in a specific “habitat”, for example the skin or mouth, intestinal tract or vagina and in recent years, increasing evidences demonstrated that human microbiota plays a crucial role in maintaining human health (1,2). Traditional cultured based methods capture only small proportion of all bacterial microbiota, but next generation sequencing DNA has closed this gap (3-5). Several data from US (HMP) or European Microbiome Project (MetaHIT) have shown, through 16S rRna analysis, that a subset of microbial genes can be found in the majority of healthy individuals, known as “core microbiome”, whereas there are variable components that may be typical of specific ethnic group, or that can be associated with geographic locations, dietary habits and diseases (3-9).
The stable healthy microbial community, which is known as colonization resistance, consistently provides protection against enteric pathogens, nutrients from food, and immune system regulation (3,4). The dynamic interactions between host and its microbiota are most clearly demonstrated in the gastrointestinal tract where the microbiota populate at a higher density. Changes in gut microbiota composition are closely related to health: this crucial defense mechanism can be impaired by several factors such as treatment with antibiotics or chemotherapy, and patients may become carriers of pathogenic bacteria, including strains that are resistant to treatment [multidrug resistant (MDR)] (10-12). The extensive use of broad-spectrum antibiotics has been linked to the spread of antimicrobial resistance in the last ten years and antimicrobial resistance is an increasing threat to human health (12).
Intestinal acquisition of resistant bacteria over the healthy microbiota is well known to be one of the major risk factor for the development of invasive infection by MDR bacteria, such as carbapenemase-producing K. pneumoniae (CP-Kp) or by Candida (13,14). Modulation of the gut microbiota might be an effective treatment strategy for restoring healthy function; however, the precise effect of antibiotics on the intestinal microbiota and the effect on the acquisition and the overgrowth of MDR bacteria needs to be further investigated.
Aim of this review is to discuss the role of gut microbiota alterations in the pathogenesis of invasive infections by MDR resistant organism and Candida by an infectious diseases perspective.
Gut microbiome
Two major phyla, Bacteroidetes and Firmicutes, dominate the healthy gut microbiota (15). Approximately 1.5 kg of bacteria lives in the human intestinal tract, and anaerobes are found at 100–1,000 time that of aerobes and facultative aerobes (2). Even if this general profile remains constant, gut microbiota exhibits both temporal and spatial shift in species distribution. From the esophagus distally to the rectum, there is a marked difference in diversity and number of bacteria ranging from 10 per gram of contents in the esophagus and stomach to 1012/gram of contents in the colon and distal gut. The large intestine constitutes over 70% of all microbes and they are generally identified by stool samples (16).
Microbiota begins to inhabit the human intestinal tract after vaginal birth, with facoltative aerobes as earliest resident, such as E. coli, Streptococci or Enterococcus spp. Normal gut microbiota has several functional aspects, such as defense against pathogens, cell proliferation and differentiation, mucus secretion, and barrier function; furthermore it has anti-inflammatory and antioxidative effects and it acts as antimicrobial protection by protecting mucosal barrier function (15). In fact, the integrity of the gut barrier is maintained by modulation of expression of proteins required for maintenance of desmosomes at epithelial villus as well as stimulation of microbial cell wall peptidoglycan or transcription of angiogenin factor 3, implicated in the development of intestinal microvasculature. The other mechanism to supervise the overgrowth of pathogenic strains is by inducing local immunoglobulins A through activation of intestinal dendritic cells (17).
During prolonged antibiotic treatments, gut microbiota can be affected by collateral damage, becoming a significant “reservoir” of MDR microorganisms with a “nosocomial profile” of antibiotic resistance. In fact, MDR bacteria that are not eliminated by antibiotic treatment, can proliferate and reach higher density in the intestinal lumen (11). In C. difficile infections (CDI) the causal role of a dysbiotic microbiota is well known, and it has been suggested that similar alterations may favor intestinal colonization by CP‐Kp (18).
Intestinal colonization by CP-Kp seems to be one of the most important risk factors for the development of CP-Kp invasive infections and selective digestive tract decontamination (SDD) of colonized patients with non-absorbed antibiotics has been suggested to reduce transmission and prevent invasive infections, although concerns are rising regarding the risk of resistance in patients who fail to respond to gut decontamination (13,19-21).
In clinical practice, colonized patients pose an epidemiological threat to other hospitalized individuals but are also in danger of developing systemic infections with gut-colonizing microorganisms, with a higher risk in the hematological setting (22).
Evidence for GI microbiome modifications
The composition of human microbiota is easily regulated by many environmental factors, particularly age, diet and antibiotic use (23). The effects of diet are well described from first year of life, since breast-fed infants are colonized by Bifidobacteria whilst bottle fed infants harbor different bacteria such as Bacteroides, Clostridium and Enterobacteriaceae (24). Diet seems to be the most important determinant in shaping the gut microbiota for the entire human life and diet habits can affect the composition of gut microbiome, causing disbiosis or in some cases protection from diseases by increasing the proportion of specific bacteria species (25). There is some evidence that gut could be colonized by organisms in utero, although the first microbiota profile is now clear that is largely shaped at delivery (26). From the age of 3, infant gut microbiota can be considered as adult microbiota since it has from 40% to 60% of similarity. Studies have shown that young children still have significant differences in proportion of Bacteroides and Bifidobacterium compared to adults. In pre-term infants, there is a predominance of Bifidobacterium and Lactobacillus that can be modified, according to the type of feeding habits (27). Although the initially developing microbiota is largely influenced by feeding, several factors during life modify gut microbiota. Gut microbiota changes according to age and its composition in the elderly differs considerably between individuals, as extensively studied in the ELDERMET cohort (28-30). In this study, a shift in the microbiota toward a Bacteroidetes-predominated population in frailer older patients compared to younger individuals was observed (28). This large variance is probably due to external factors influencing the microbiota, such as diet, exercise and inactivity, and medication (29). Several ongoing studies, such as the NuAge project, will apply multi-omics analysis to determine if microbiota changes are related to or are the cause of health loss and how other physiological processes might influence these changes (30). This will establish any possible impact on aging health status, by or even modulating the microbiota composition and function, the possibility to target microbiota for interventions to promote healthier aging (30,31).
A strong body of evidence has now demonstrated that the use of antibiotics has long-term effects on the ecology of gut microbiota, with a reduction of taxonomic diversity and persistence of changes in the majority of individuals. Data showed that the effect of short-term use of broad-spectrum antibiotics with predominant anaerobic coverage could last up to 2 years, with a persistent depletion of the diversity of Bacteroides (32,33). Different studies have demonstrated that a short course of H. pylori eradication results in a dramatic reduction in the diversity of Actinobacteria that may endure over 4-year period, as well as it may lead to an overgrowth of potentially pathogenic organism including MDR Enterobacteriaceae and yeast (34,35). These data were confirmed in recent studies using molecular techniques based on analysis of 16S rRNA, in which the loss of diversity and changes in different bacteria taxa have been reported in patients treated with prolonged antibiotic therapies (36). Moreover, the antibiotic-induced alterations increase susceptibility to gastrointestinal infections, such as salmonellosis and diarrhea and colitis by C. difficile (37). Thus, one of the major concerns in addition to the alteration of the normal microbiota diversity, is that the use of broad spectrum antibiotics can lead to gut colonization by MDR strain of bacteria that could easily overwhelm the bloodstream (13,38).
“Enteropathogenetic Syndromes”
Dysbiosis is represented by an alteration and ‘‘imbalance” of the microbiome without any chance to physiological recovery, due to several factors. The causal role of dysbiosis is well known for CDI, in which the altered microbiome creates a favourable setting for C. difficile colonization, sporulation, spore germination, and toxin production. Dysbiotic state is maintained and promoted by C. difficile disruption of epithelial barrier function and alterations of inflammatory responses, and only an intervention targeted to restore a physiological microbiome is helpful to clear CDI (39). Thus, similar alteration may be favoring colonization of MDR bacteria such as carbapenemase producing CP-Kp or ESBL or an excessive intestinal growth of Candida spp, favoring bloodstream infections (40). There are some reports of candidemia following CDI, of CP-Kp bloodstream infections (BSI) associated with candidemia, of early infections by CP-Kp, and of possible interactions amongst antibiotics use to treat CDI and Candida colonization (40-43). Prescott et al. showed that in patients with previous CDI, there is a higher risk of sepsis in the 90 days after admission, related to the persistence of altered microbiota (41). Interestingly, in in vivo models of gastrointestinal candidiasis Cole et al. evaluated the role of gastrointestinal mucosa colonization, alterations of the integrity of the mucosal epithelium and damage of mucosal immunity in the development of invasive candidiasis (14).
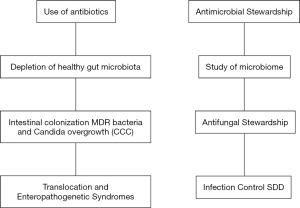
Nowadays, it is common knowledge that the gastrointestinal tract has the crucial role of main reservoir for human diseases by Candida spp. and for epidemic spread of MDR bacteria such as CP-Kp and C. difficile. For these reasons, we proposed a new acronyme “CCC” to highlight the similar pathogenetic pathway of Candida spp, C. difficile and CP-Kp infections (38). In order to stress the importance of gastrointestinal alterations in promoting intestinal colonization, overgrowth and diseases by these pathogens, we proposed the term “enteropathogenetic syndromes”, to describe enteropathogenetic endogenous opportunistic infections caused by “CCC” (18). To reduce the opportunity of enteropathogenetic syndromes, there is a strong need for new antimicrobial and antifungal stewardship approaches, aiming to reduce antimicrobial consumption, thus protecting and maintaining the gut microbiota (44) (Figure 1).
How to eradicate MDR colonization by CP-Kp
SDD
Along with infection control measures such as patient isolation or cohorting with dedicated staff, SDD with nonabsorbed antibiotics has been proposed as an effective strategy to reduce transmission and preventing subsequent infectious episodes in colonized patients by CP-Kp. One of the most used regimen for SDD is oral gentamicin or combination of gentamicin plus polymyxin E (45). The efficacy of SDD with gentamicin 80 mg four times daily was evaluated by Tascini et al. in 50 patients colonized by gentamicin-susceptible CP-Kp, with or without concomitant systemic antibiotic therapy (19). The overall decontamination rate was 68% and at the 6-month follow-up, a CP-Kp infection was observed only in 15% of successfully decontaminated patients compared to 73% of persistent carriers (19).
In our center, we proposed a multiple-step intervention strategy in hematological patients undergoing allogenic bone narrow transplantation (HSCT) colonized by CP-Kp. Steps included oral gentamicin within 20 days before HSCT, treatment of febrile neutropenia with intravenous tigecycline 100 mg bid and piperacillin-tazobactam at standard dosages and early appropriate combination therapy for patients with severe sepsis. In our small series all patients survived, no resistance to oral gentamicin was observed and 60% of patients had negative rectal swabs after transplant (21).
Lübbert et al. described 14 patients colonized by CP-Kp, treated with 7 days of combination of colistin and oral gentamicin, and applying colistin/gentamicin gel (0.5 g) to the oral cavity. Decolonisation of CP-Kp was achieved in 43% of patients treated with the antibiotics, but there was an increased risk of secondary resistance to colistin and gentamicin following SDD (19% and 45% increase, respectively) (46). Thus, the risk of resistance should be evaluated before starting SDD, since oral topic antibiotic therapy can be useful but may favor the emergence of resistant CP-Kp, especially in patients with failures of decontamination regimens.
Fecal microbiota transplantation (FMT)
A large body of evidence, including randomised controlled trials (RCTs), systematic reviews and meta-analyses, proved clear evidence that FMT is a highly effective treatment for CDI and based on these data both the European Society for Microbiology and Infectious Disease and the American College of Gastroenterology recommended FMT as a treatment for recurrent CDI (47-55). Promising findings suggest that FMT may play a role also in the management of other disorders associated with the alteration of gut microbiota, such as to decolonize patients from MDR bacteria. So far only case reports have been reported using FMT to decolonize from MDR bacteria, but six trials are ongoing (56-61). Regarding the safety of this procedure, no serious adverse events were reported in any of the case reports published to date (56). However, long term monitoring of the efficacy of FMT still need to be further investigated, since current evidence suggest a temporary more than permanent decolonization from MDR bacteria (62).
Conclusions: targeting microbiota
Variability of human microbiota is closely related to health and diseases. There are many ways to regulate the composition of gut microbiota, therefore targeting the gut microbiota has been proposed as a new therapeutic approach to tackle several diseases (23). FMT for recurrent CDI is a major example of a successful approach to restore microbiota complexity, with success rates of approximately 90% (47-53). Regarding antimicrobial resistance and the battle against MDR bacteria, intestinal microbiota could be the key to prevent or eradicate MDR colonization. So far, there are no data describing which bacterial species provide the best protection against MDR colonization, thus the most promising approach might be FMT (62). So far, there are several obstacles to determine the role of microbiota in human health, first of all samples collection and methods of analysis (23). There are several methods for determining the biodiversity and 16S rRNA sequencing is the most used, although several reports have shown that different amplification strategies or sample collections were associated with different results (63,64). Shotgun whole genome sequencing is usually performed to obtain information about the composition of the microbiome and association with metabolic patterns with diseases, but standardized samples collection and analysis are important steps for successful research on microbiota. Second, microbiota is affected by dietary habits and different ethnic groups are different, highlighting another complexity for clinical studies (65). Third, integration of omics data with phenotypic data of the diseases is important. Moreover, few data are available so far, and the interaction of microbioma with human immunogenetic determinants, such as proteins and genes (66).
In conclusion, there are some basic questions that still need to be answered to understand the gut microbioma complexities and therapeutics perspectives. As we progress from an era of broad spectrum antibiotics to a more effective stewardship approaches, the knowledge of microbiome and its functions will help to individualized medicine, either preventive or curative (3,44,67). In the future targeting or restoring microbiome may represent an effective and complementary therapeutic strategy.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aoi.2017.08.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev 2012;70:S45-56. [Crossref] [PubMed]
- Warner BB, Hamvas A. Lungs, microbes and the developing neonate. Neonatology 2015;107:337-43. [Crossref] [PubMed]
- Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology 2009;136:2015-31. [Crossref] [PubMed]
- Martínez I, Muller CE, Walter J. Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS ONE 2013;8:e69621 [Crossref] [PubMed]
- Gibson MK, Pesesky MW, Dantas G. The yin and yang of bacterial resilience in the human gut microbiota. J Mol Biol 2014;426:3866-76. [Crossref] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. [Crossref] [PubMed]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59-65. [Crossref] [PubMed]
- Ursell LK, Metcalf JL, Parfrey LW, et al. Defining the Human Microbiome. Nutr Rev 2012;70:S38-44. [Crossref] [PubMed]
- Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature 2007;449:804-10. [Crossref] [PubMed]
- Montassier E, Gastinne T, Vangay P, et al. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment Pharmacol Ther 2015;42:515-28. [Crossref] [PubMed]
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 2013;13:790-801. [Crossref] [PubMed]
- Becattini S, Taur Y, Pamer EG. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends Mol Med 2016;22:458-78. [Crossref] [PubMed]
- Tumbarello M, Trecarichi EM, Tumietto F, et al. Predictive models for identification of hospitalized patients harboring KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2014;58:3514-20. [Crossref] [PubMed]
- Cole GT, Halawa AA, Anaissie EJ. The role of the gastrointestinal tract in hematogenous candidiasis: from the laboratory to the bedside. Clin Infect Dis 1996;22:S73-88. [Crossref] [PubMed]
- Jandhyala SM, Talukdar R, Subramanyam C, et al. Role of the normal gut microbiota. World J Gastroenterol 2015;21:8787-803. [Crossref] [PubMed]
- Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220-30. [Crossref] [PubMed]
- He B, Xu W, Santini PA, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity 2007;26:812-26. [Crossref] [PubMed]
- De Rosa FG, Corcione S, Raviolo S, et al. Candidemia, and infections by Clostridium difficile and carbapenemase-producing Enterobacteriaceae: new enteropathogenetic opportunistic syndromes? Infez Med 2015;23:105-16. [PubMed]
- Tascini C, Sbrana F, Flammini S, et al. Oral gentamicin gut decontamination for prevention of KPC-producing Klebsiella pneumoniae infections: relevance of concomitant systemic antibiotic therapy. Antimicrob Agents Chemother 2014;58:1972-6. [Crossref] [PubMed]
- Zuckerman T, Benyamini N, Sprecher H, et al. SCT in patients with carbapenem resistant Klebsiella pneumoniae: a single center experience with oral gentamicin for the eradication of carrier state. Bone Marrow Transplant 2011;46:1226-30. [Crossref] [PubMed]
- De Rosa FG, Corcione S, Raviolo S, et al. Management of carbapenem-resistant K. pneumoniae in allogenic stem cell transplant recipients: the Turin bundle. New Microbiol 2017;40:143-5. [PubMed]
- Bilinski J, Grzesiowski P, Sorensen N, et al. Fecal Microbiota Transplantation in Patients With Blood Disorders Inhibits Gut Colonization With Antibiotic-Resistant Bacteria: Results of a Prospective Single-Center Study. Clin Infect Dis 2017;65:364-70. [Crossref] [PubMed]
- Bi Y, Qin N, Yang R. Human Microbiota: A Neglected “Organ” in Precision Medicine. Inf Dis Transl Med 2015;1:63-72.
- Martín R, Jiménez E, Heilig H, et al. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol 2009;75:965-9. [Crossref] [PubMed]
- Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 2013;110:9066-71. [Crossref] [PubMed]
- Moya-Pérez A, Luczynski P, Renes IB, et al. Intervention strategies for cesarean section-induce alterations in the microbiota-gut-brain axis. Nutr Rev 2017;75:225-40. [Crossref] [PubMed]
- Fanaro S, Chierici R, Guerrini P, et al. Intestinal Microflora in early infancy: composition and development. Acta Paediatr Suppl 2003;91:48-55. [PubMed]
- Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 2011;108:4586-91. [Crossref] [PubMed]
- Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178-84. [PubMed]
- Santoro A, Pini E, Scurti M, et al. Combating inflammaging through a Mediterranean whole diet approach: the NU-AGE project's conceptual framework and design. Mech Ageing Dev 2014;136-137:3-13. [Crossref] [PubMed]
- O'Toole PW, Jeffery IB. Gut microbiota and aging. Science 2015;350:1214-5. [Crossref] [PubMed]
- Jernberg C, Löfmark S, Edlund C, et al. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 2007;1:56-66. [Crossref] [PubMed]
- Dethlefsen L, Huse S, Sogin ML, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. Plos Biol 2008;6:e280 [Crossref] [PubMed]
- Jakobsson HE, Jernberg C, Andersson AF, et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 2010;5:e9836 [Crossref] [PubMed]
- Adamsson I, Nord CE, Lundquist P, et al. Comparative effects of omeprazole, amoxycillin plus metronidazole versus omeprazole, clarithromycin plus metronidazole on the oral, gastric and intestinal microflora in Helicobacter pylori-infected patients. J Antimicrob Chemother 1999;44:629-40. [Crossref] [PubMed]
- Koh AY. Potential for Monitoring Gut Microbiota for Diagnosing Infections and Graft-versus-Host Disease in Cancer and Stem Cell Transplant Patients. Clin Chem 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Bartlett JG. Clinical Practice antibiotic associated diarrhea. N Engl J Med 2002;346:334-9. [Crossref] [PubMed]
- De Rosa FG, Corcione S, Pagani N, et al. From ESKAPE to ESCAPE, from KPC to CCC. Clin Infect Dis 2015;60:1289-90. [Crossref] [PubMed]
- Almeida R, Gerbaba T, Petrof EO. Recurrent Clostridium difficile infection and the microbiome. J Gastroenterol 2016;51:1-10. [Crossref] [PubMed]
- Nerandzic MM, Mullane K, Miller MA, et al. Reduced acquisition and overgrowth of vancomycin-resistant enterococci and Candida species in patients treated with fidaxomicin versus vancomycin for Clostridium difficile infection. Clin Infect Dis 2012;55:S121-6. [Crossref] [PubMed]
- Prescott HC, Dickson RP, Rogers MA, et al. Hospitalization Type and Subsequent Severe Sepsis. Am J Respir Crit Care Med 2015;192:581-8. [Crossref] [PubMed]
- Guastalegname M, Russo A, Falcone M, et al. Candidemia subsequent to severe infection due to Clostridium difficile: is there a link? Clin Infect Dis 2013;57:772-4. [Crossref] [PubMed]
- Papadimitriou-Olivgeris M, Spiliopoulou A, Fligou F, et al. Association of KPC-producing Klebsiella pneumoniae colonization or infection with Candida isolation and selection of non-albicans species. Diagn Microbiol Infect Dis 2014;80:227-32. [Crossref] [PubMed]
- Corcione S, Pagani N, Forni N, et al. Start Smart With Antimicrobial Stewardship Clin Infect Dis 2015;61:1033-4. [Crossref] [PubMed]
- Saidel-Odes L, Polachek H, Peled N, et al. A randomized, double-blind, placebo-controlled trial of selective digestive decontamination using oral gentamicin and oral polymyxin E for eradication of carbapenem-resistant Klebsiella pneumoniae carriage. Infect Control Hosp Epidemiol 2012;33:14-9. [Crossref] [PubMed]
- Lübbert C, Faucheux S, Becker-Rux D, et al. Rapid emergence of secondary resistance to gentamicin and colistin following selective digestive decontamination in patients with KPC-2-producing Klebsiella pneumoniae: a single-centre experience. Int J Antimicrob Agents 2013;42:565-70. [Crossref] [PubMed]
- van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013;368:407-15. [Crossref] [PubMed]
- Cammarota G, Masucci L, Ianiro G, et al. Randomised clinical trial: faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection Aliment Pharmacol Ther 2015;41:835-43. [Crossref] [PubMed]
- Lee CH, Steiner T, Petrof EO, et al. Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: a randomized clinical trial. JAMA 2016;315:142-9. [Crossref] [PubMed]
- Kassam Z, Lee CH, Yuan Y, et al. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol 2013;108:500-8. [Crossref] [PubMed]
- Cammarota G, Ianiro G, Gasbarrini A. Fecal microbiota transplantation for the treatment of Clostridium difficile infection: a systematic review. J Clin Gastroenterol 2014;48:693-702. [Crossref] [PubMed]
- Drekonja D, Reich J, Gezahegn S, et al. Fecal microbiota transplantation for Clostridium difficile infection: a systematic review. Ann Intern Med 2015;162:630-8. [Crossref] [PubMed]
- Mattila E, Uusitalo-Seppälä R, Wuorela M, et al. Fecal transplantation, through colonoscopy, is effective therapy for recurrent Clostridium difficile infection. Gastroenterology 2012;142:490-6. [Crossref] [PubMed]
- Surawicz CM, Brandt LJ, Binion DG, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol 2013;108:478-98. [Crossref] [PubMed]
- Cammarota G, Ianiro G, Tilg HEuropean FMT Working Group, et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017;66:569-80. [Crossref] [PubMed]
- Manges AR, Steiner TS, Wright AJ. Fecal microbiota transplantation for the intestinal decolonization of extensively antimicrobial-resistant opportunistic pathogens: a review. Infect Dis (Lond) 2016;48:587-92. [Crossref] [PubMed]
- Stool transplantation to reduce antibiotic resistance transmission. Accessed 10 July 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT02461199
- Fecal transplant for MDR pathogen decolonization. Accessed 10 July 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT02906774
- FMT for multidrug resistant organism reversal. Accessed 10 July 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT02312986
- Biotherapy for MRSA enterocolitis. Accessed 10 July 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT02390622
- FMT for MDRO colonization after infection in renal transplant recipients. Accessed 10 July 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT02922816
- Bassetti M, Poulakou G, Ruppe E, et al. Antimicrobioal resistance in the next 30 years, humankind, bugs and drugs: a visionary approach. Intensive Care Med 2017; [Epub ahead of print]. [Crossref]
- Walker AW, Martin JC, Scott P, et al. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015;3:26. [Crossref] [PubMed]
- Wagner Mackenzie B, Waite DW, et al. Evaluating variation in human gut microbiota profiles due to DNA extraction method and inter-subject differences. Front Microbiol 2015;6:130. [Crossref] [PubMed]
- Zhang J, Guo Z, Xue Z, et al. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J 2015;9:1979-90. [Crossref] [PubMed]
- Guo X, Liang Y, Zhang Y, et al. Innate Lymphoid Cells Control Early Colonization Resistance against Intestinal Pathogens through ID2-Dependent Regulation of the Microbiota. Immunity 2015;42:731-43. [Crossref] [PubMed]
- Baietto L, Corcione S, Pacini G, et al. A 30-years review on pharmacokinetics of antibiotics: is the right time for pharmacogenetics? Curr Drug Metab 2014;15:581-98. [Crossref] [PubMed]
Cite this article as: Corcione S, De Rosa FG. The gastrointestinal microbiome and the enteropathogenetic syndromes: an infectious diseases perspective. Ann Infect 2017;1:4.


